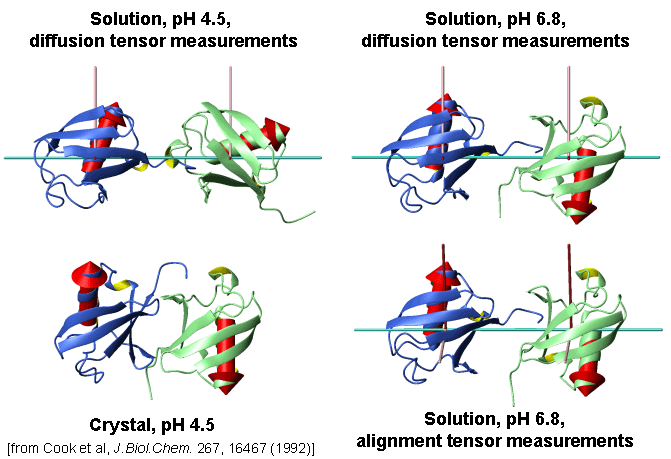
Conformations of Lys48-linked di-Ubiquitin under various conditions
[Varadan et al., J. Mol. Biol. 324 (2002) 637-647]
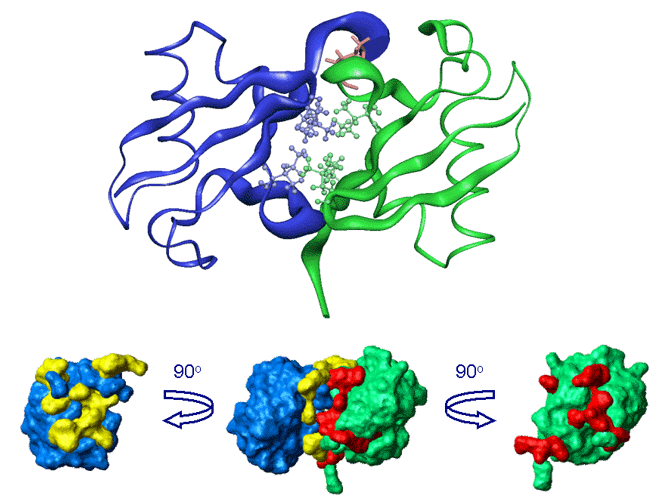
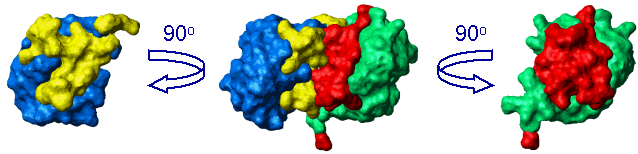
Interdomain interface in Lys48-linked di-Ubiquitin at pH6.8 mapped by chemical shift perturbations
[Varadan et al., J. Mol. Biol. 324 (2002) 637-647]
TEMPO-accessibility data: Interdomain interface in Lys48-linked di-Ubiquitin is not locked.
[Varadan et al., J. Mol. Biol. 324 (2002) 637-647]
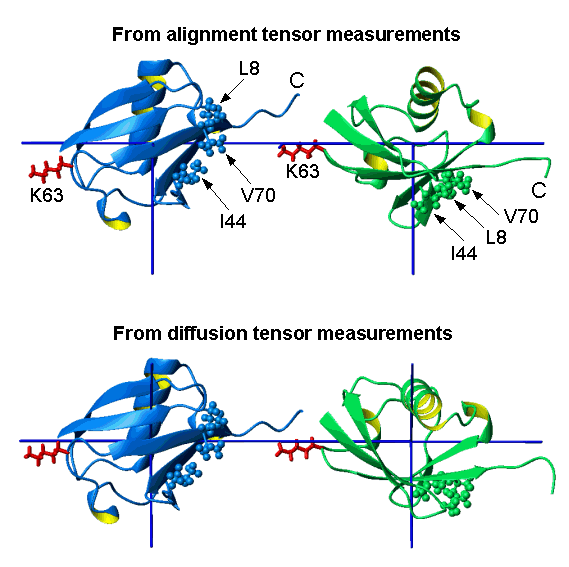
Conformation of Lys63-linked di-Ubiquitin
[Varadan et al., J. Biol. Chem. 279 (2004) 7055-7063]
Alternatively linked polyubiquitin chains adopt distinct conformations.
[Varadan et al., J. Biol. Chem. 279 (2004) 7055-7063]
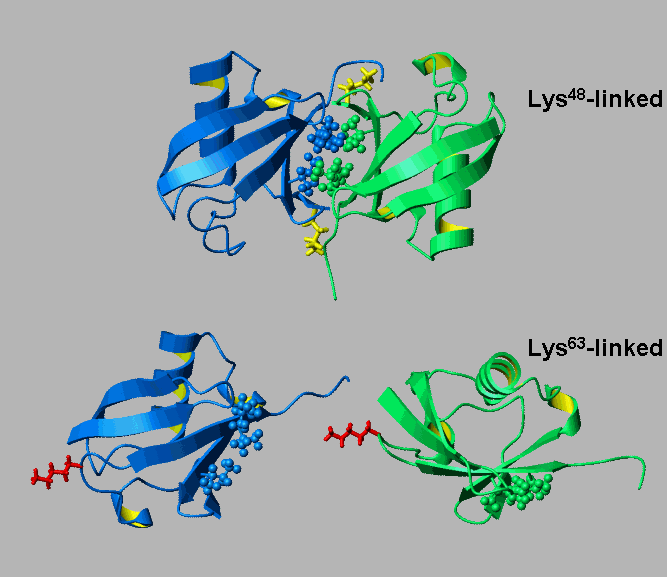
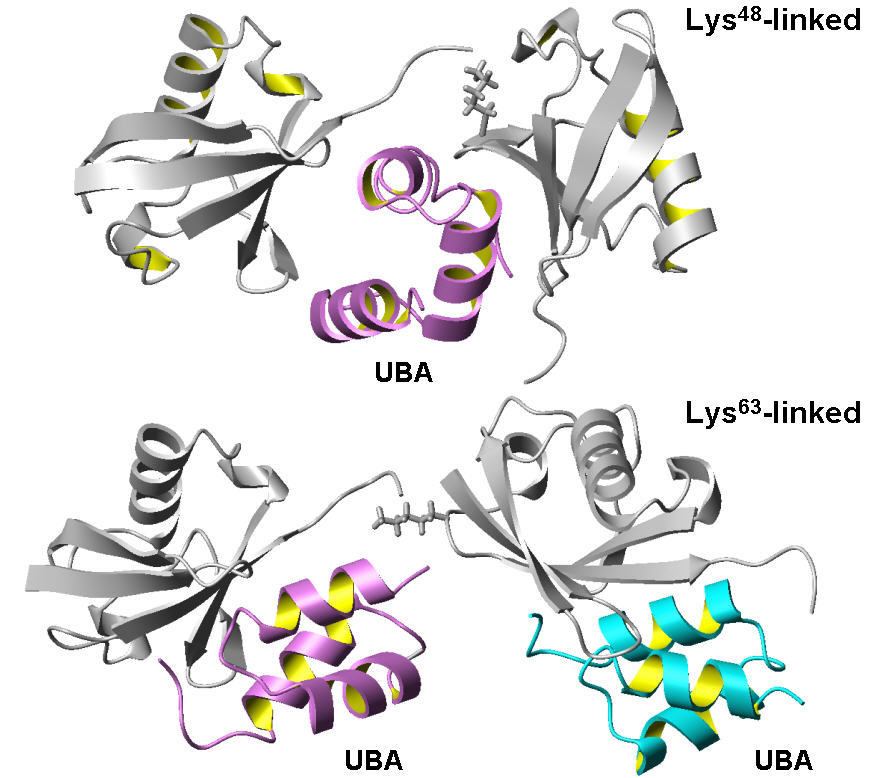
Alternatively linked polyubiquitin chains could present distinct recognition signals.
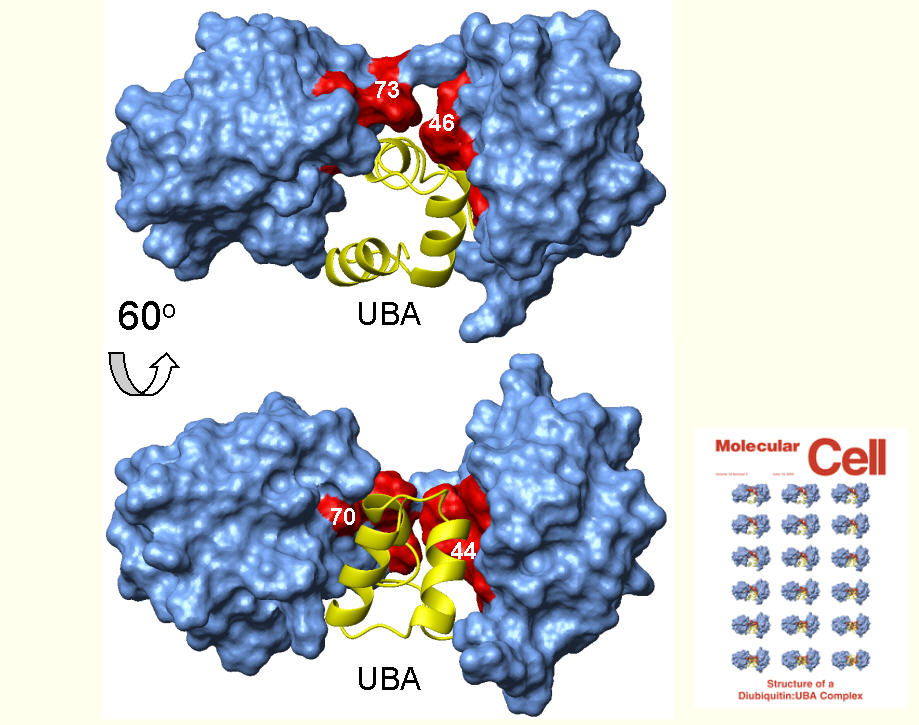
Binding of the UBA2 domain from hHR23A to Lys48- and Lys63-linked di-Ubiquitin chains
[Varadan et al., Molecular Cell 18 (2005) 687-698; J. Biol. Chem. 279 (2004) 7055-7063.]
Complex of Lys48-linked di-Ubiquitin with the UBA2 domain from hHR23A. (PDB code: 1zo6)
[Varadan et al., Molecular Cell 18 (2005) 687-698]
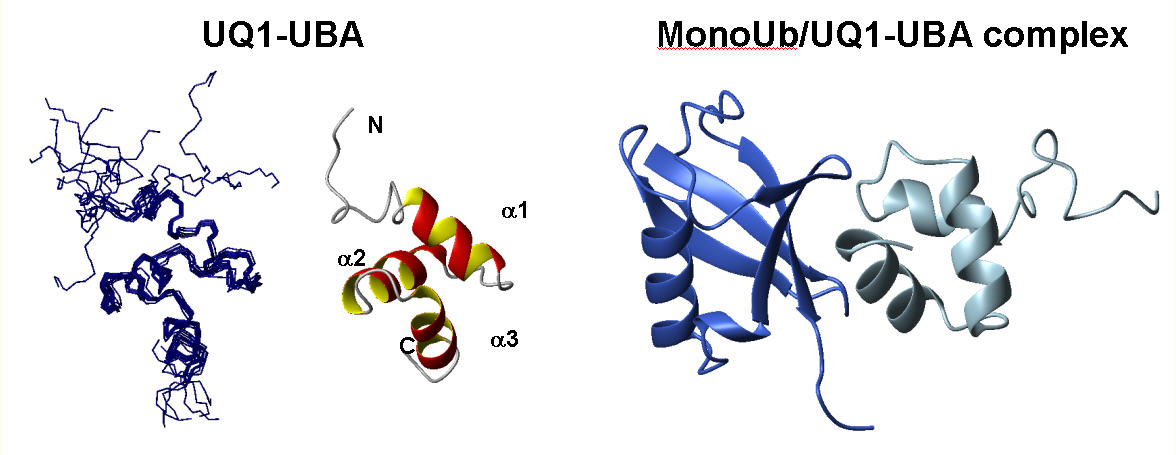
Solution structure of UBA domain of Ubiquilin-1 (hPLIC-1) and its complex with Ubiquitin. (PDB code: 2jy6)
[Zhang et al., J. Mol. Biol. (2008) in press. Web publ. Dec 23, 2007]
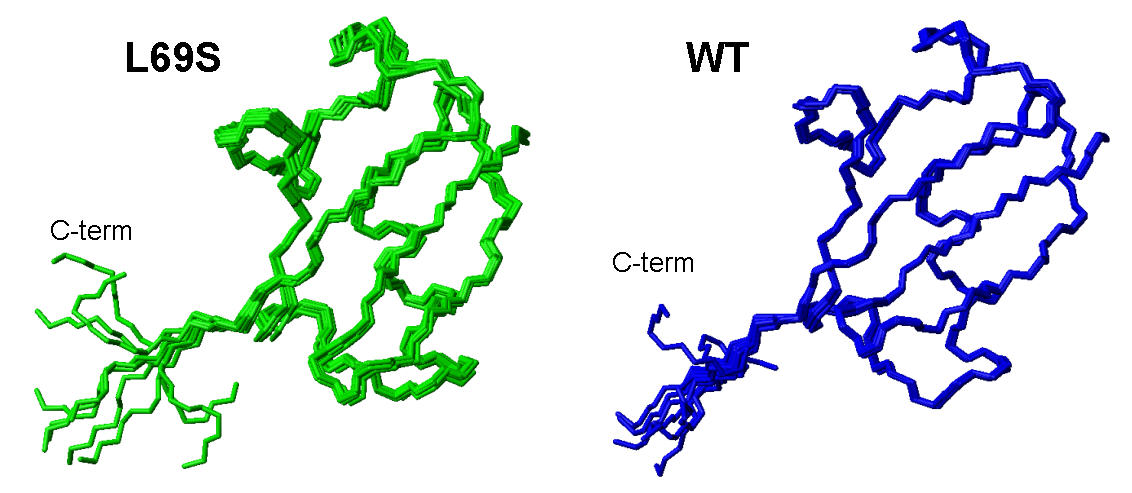
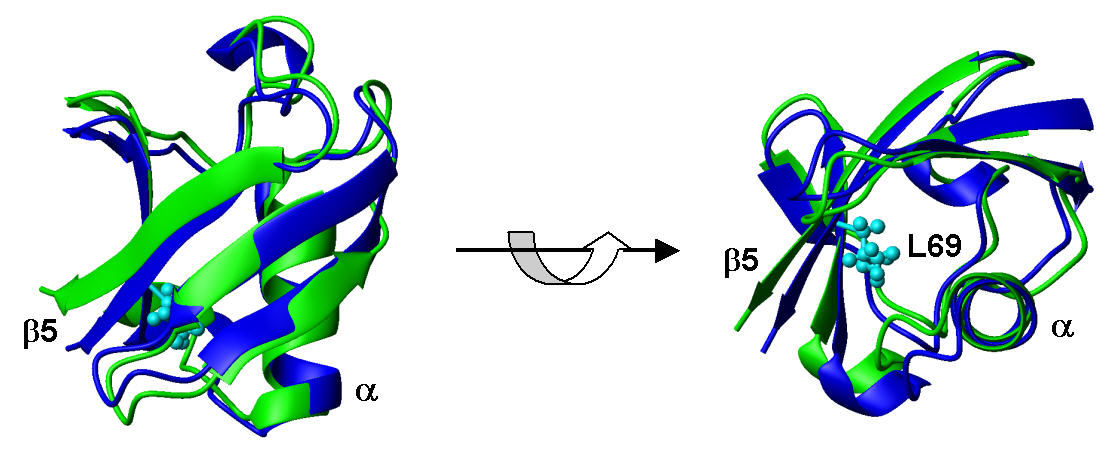
Solution structure of L69S mutant of yeast Ubiquitin (PDB code: 2jwz) and its comparison with wild type Ubiquitin
[Haririnia et al., J. Mol. Biol. 375 (2008) 979-996]
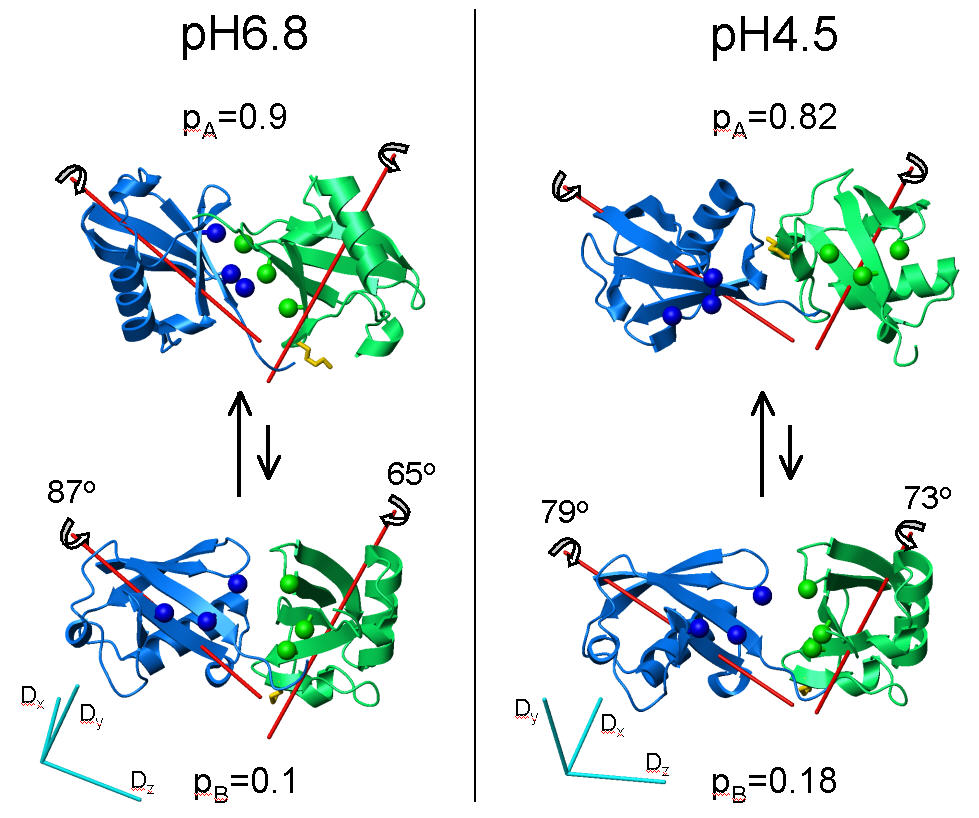
Interdomain dynamics in Lys48-linked di-Ubiquitin at various pH conditions.
[Y. Ryabov & D. Fushman, Proteins 63 (2006) 787-796; J. Am. Chem. Soc. 129 (2007) 3515-3527]
_______________________________________________________________
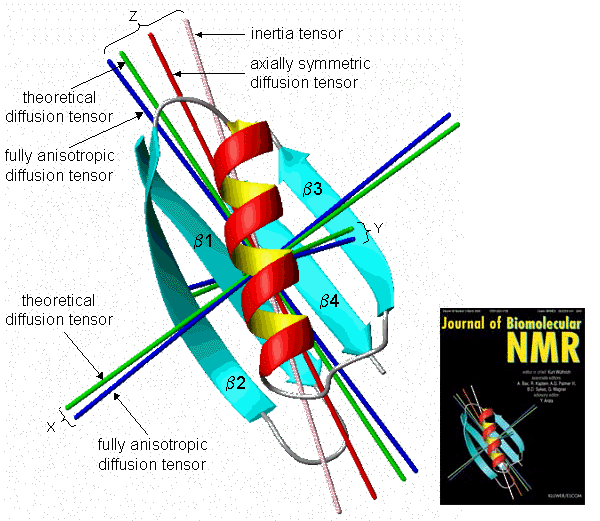
Experimental and theoretical axes of the rotational diffusion tensor in the B3 domain of protein G
[J.B.Hall & D.Fushman, J.Biomol.NMR 27 (2003) 261-275]